Research
Pathogenic Escherichia coli (E. coli) causing diarrhoea kills over half a million children under five in low and middle-income countries each year. E. coli is also a leading cause of community- and hospital-acquired bloodstream infections and a major problem in the animal industry. Adhesins play a key role in bacterial pathogenesis, enabling the bacteria to colonise and cause infection, but are also involved in survival in secondary environments, like water. Furthermore, adhesins’ ability to adhere to cells determines tissue and host specificity. A deep understanding of the pathogen’s transmission dynamics, how they evolve, how they interact with hosts, and the distribution and diversity of the virulome (set of genes that contribute to the virulence of a bacterium) is of great significance for vaccine development.
In light of these realities, the goal of the research in the von Mentzer Lab is to unravel the transmission dynamics of isolates between different hosts and the transmission of genetic elements that drive virulence and resistance in bacteria. To do this, we use data-driven genomics for high-throughput and large-scale analysis, followed by experimental corroboration when needed. Previous research by Astrid von Mentzer centred around the major diarrhoea-causing enterotoxigenic Escherichia coli (ETEC) bacteria provides the cornerstones from which we will focus on the evolution and emergence of pathogenic bacteria, with a link to how vaccines can confer broad protection against different bacterial variants and identify the drivers behind their emergence in humans and animals.
Funded by the Alice and Knut Wallenberg Foundation, Research Council and Sahlgrenska Academy, our research group combines computational and experimental approaches to understand the evolution of E. coli pathotypes and hybrid pathotypes, as well as investigating the evolution and diversity of antibiotic resistance and virulence genes and their species dynamics. We are also part of the Data-Driven Life Science (DDLS) consortium, which enables us to collaborate with leading experts and leverage cutting-edge technologies in our research.
Our main focus areas are:
-
Applying large-scale genomics and metagenomics to investigate the evolution of pathogenic E. coli. This involves analysing vast genomic, clinical, and environmental datasets to uncover how these bacteria spread and evolve into new, potentially hyper-virulent and hyper-resistant strains.
-
Using a ‘One Health’ approach to study the transmission of enterotoxigenic E. coli across ecological niches. This approach integrates data from humans, animals, and the environment to understand the zoonotic potential and cross-host transmission dynamics of pathogenic E. coli.
-
Combining genomic predictions with experimental methodologies to validate findings. This includes using advanced computational methods, such as phylogenetic inference and pangenome analyses, to analyse large-scale data and develop models that predict high-risk genetic elements and emerging pathotypes.
Additionally, we are expanding our research to incorporate machine learning and artificial intelligence (AI) to enhance our understanding of the evolution of pathogenic E. coli. By leveraging these advanced computational tools, we aim to identify patterns and predict the emergence of virulent and resistant strains more accurately, thus contributing to more effective vaccine development and public health strategies.
The impact of our research includes:
- Identifying high-risk genetic elements to enhance diagnostics and vaccine development.
- Predicting high-risk clades for early detection and containment of zoonotic transmissions.
- Detecting emerging bacterial pathotypes and assessing their disease potential to proactively develop effective diagnostics and vaccines.
Our goal over the next five years is to build an interdisciplinary team comprising experts in infection biology, data science, machine learning, epidemiology, and microbial genomics. This team is committed to addressing fundamental gaps in knowledge related to the spread, evolution, and emergence of pathogenic bacteria. Through our collaborative efforts with the DDLS consortium and other international partners, we aim to develop One Health interventions that can mitigate the impact of these bacteria on global public health.
Highlighted
All
2024

2023
2022
2021
2020
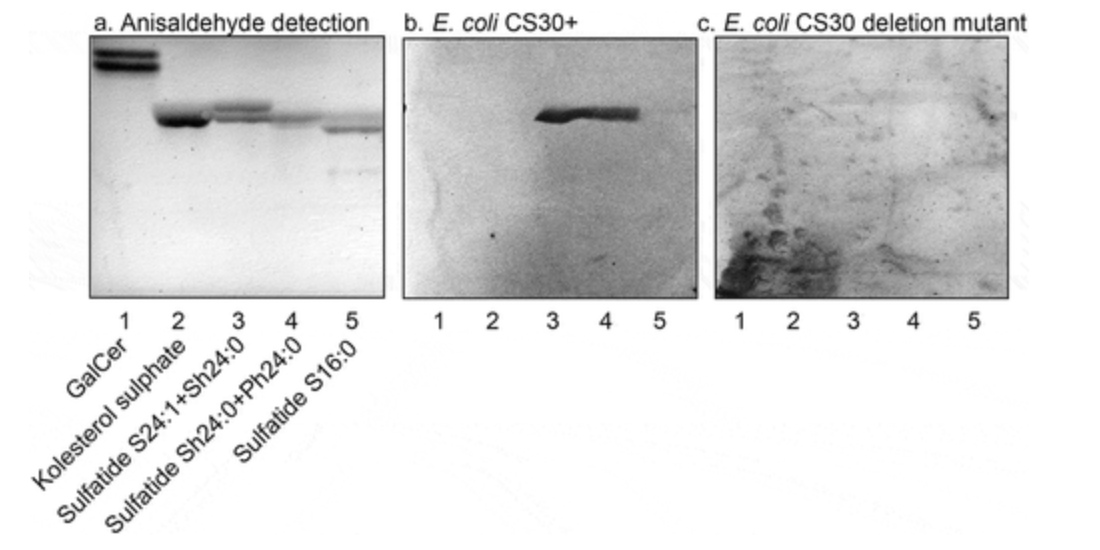
2018

2017
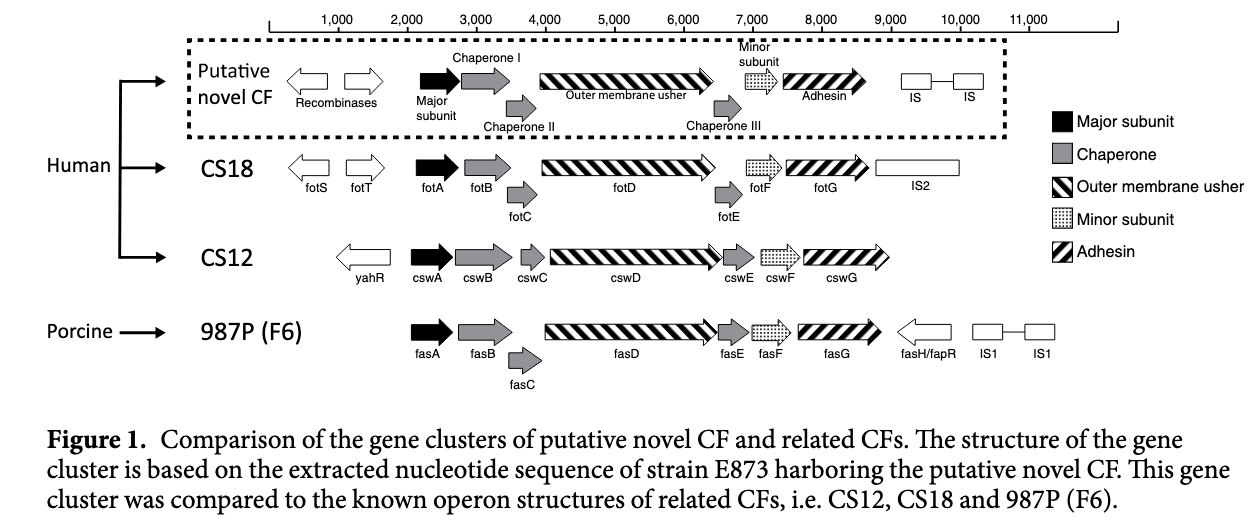
2016
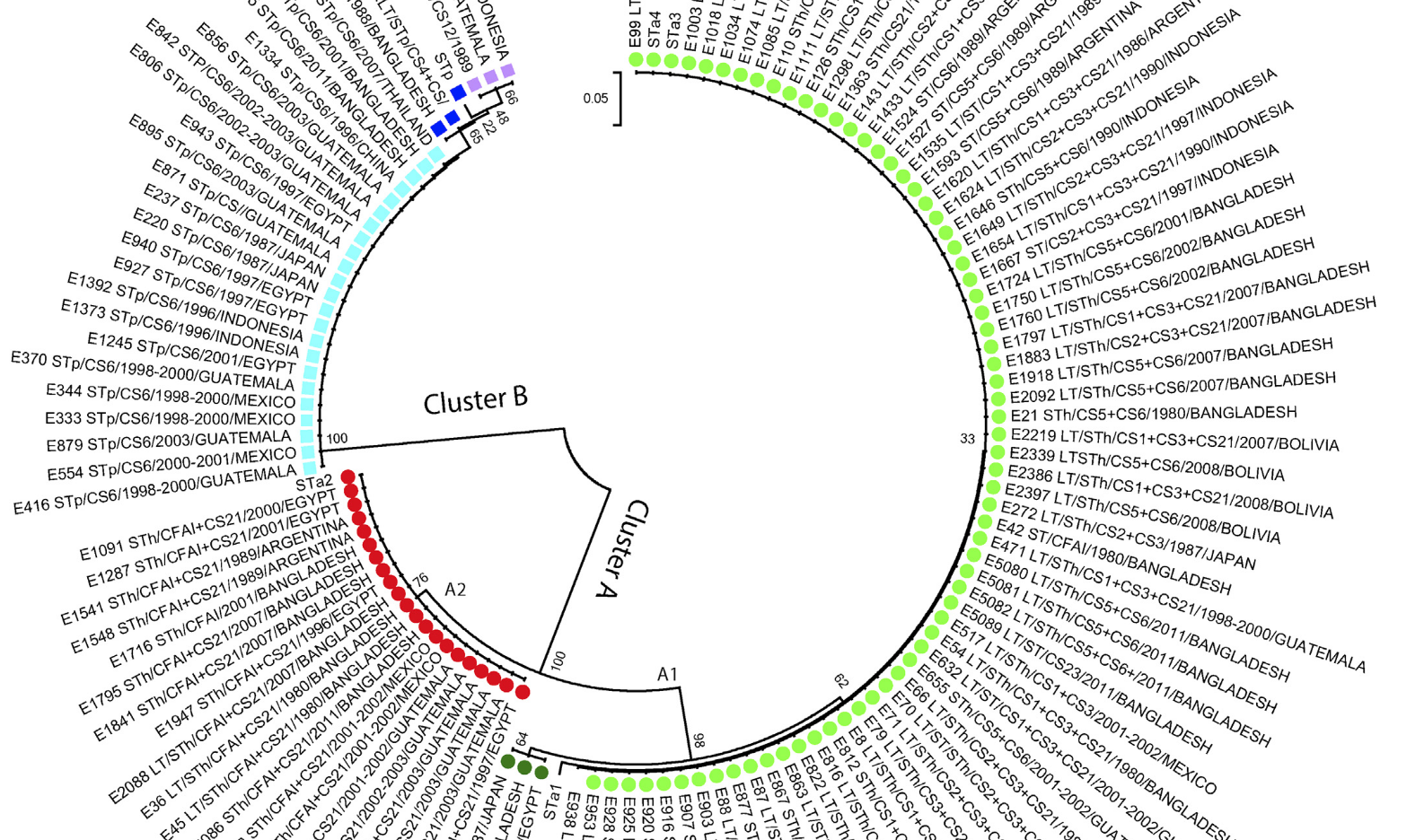
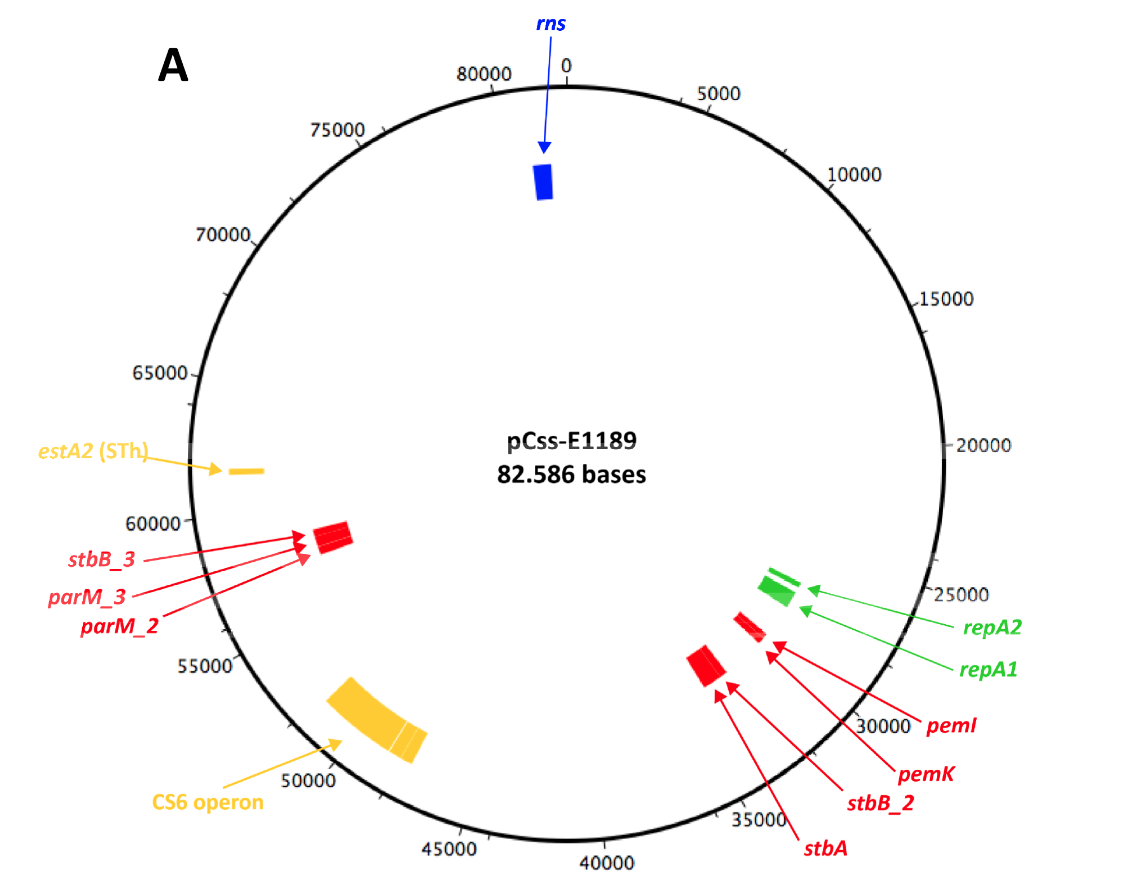
2015
2014
